As a instructor I offer a class, in which I let master students choose a topic for practical work during a semester. I usually give them a rough description, which included last time a raspberry pi, a camera, and a neural network.
Some students have chosen to work on fruit recognition with a camera. So the scenario is the following: The camera is connected to a raspberry pi. The camera observes a clean table. As soon as a user puts a fruit onto the table, the user can hit a button on a shield attached on the raspberry pi. The button triggers the camera to take an image. Then, the image is fed into a trained neural network for image categorization. The category was then fed into a speech synthesizer to speak out the category.
The type of neural network my students and I used is a multi-categorical neuronal network. So the goal was to feed the neuronal network with image and a category will come out as an output.
Preparing the Data
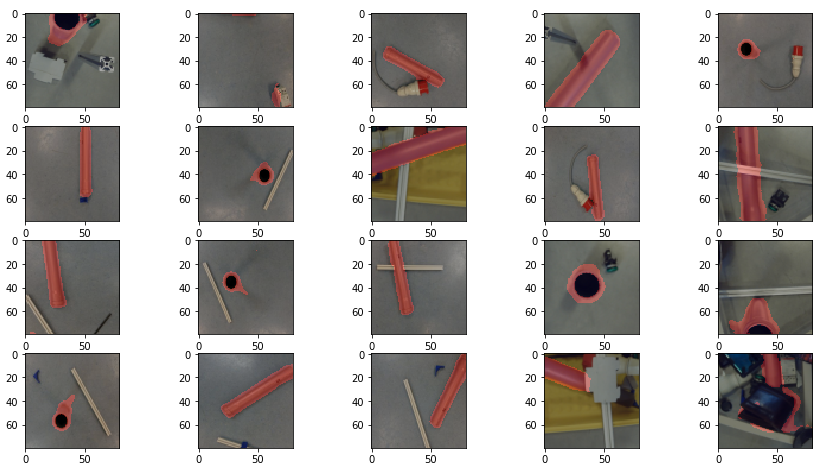
In the beginning we chose fruit images from a database which is available on github. You find it here. It had about 120 different categories of fruits and vegetables available. The problem we find with these images are, that the fruits and vegetables seemed to be perfect looking which is in reality not the case. The variation of fruit images within one category also seemed to be very limited. On the one hand, they do have many images within each category, on the other hand it looks like each image from one category only comes from a perfect fruit photographed in different positions.
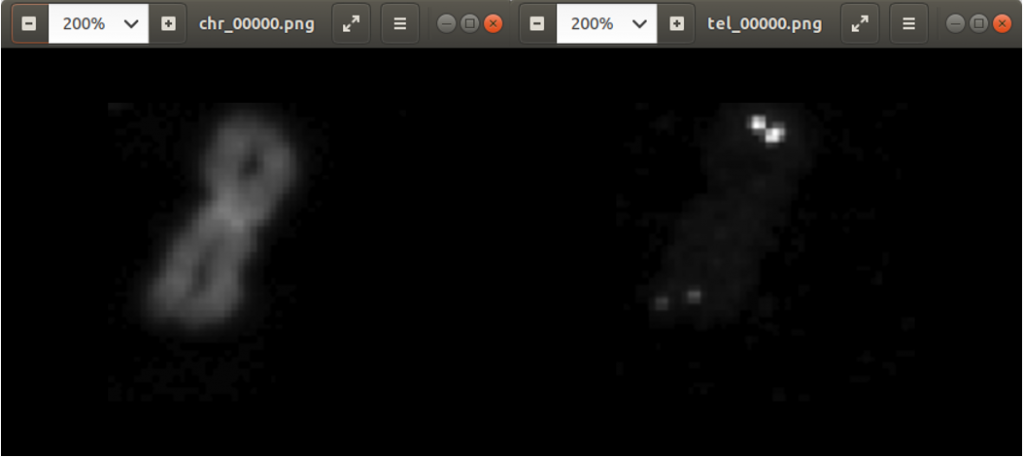
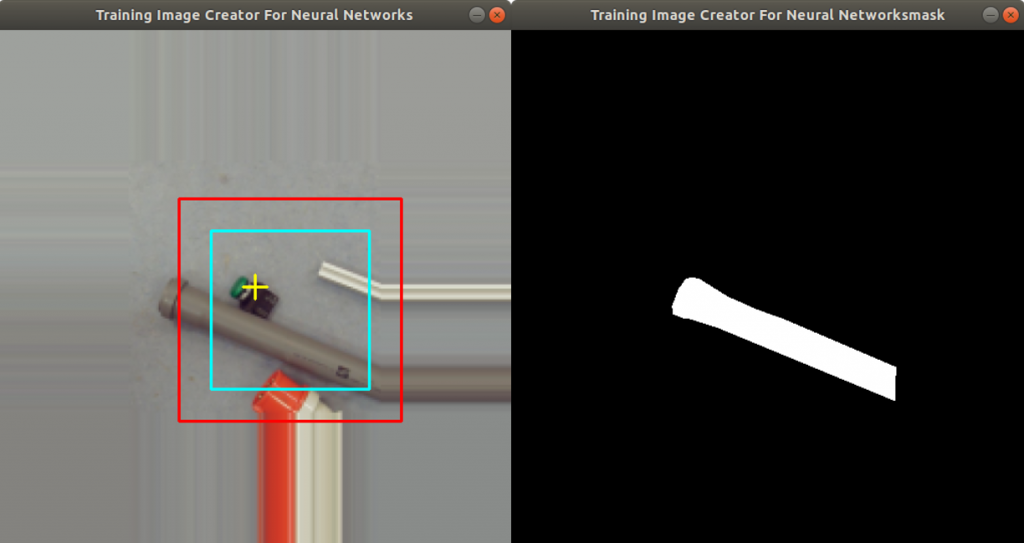
The fruits fill out the complete image, as well. When you photograph a fruit from a table, this is in general not the case. The left part of Figure 1 shows an orange which fills in only part of the image.
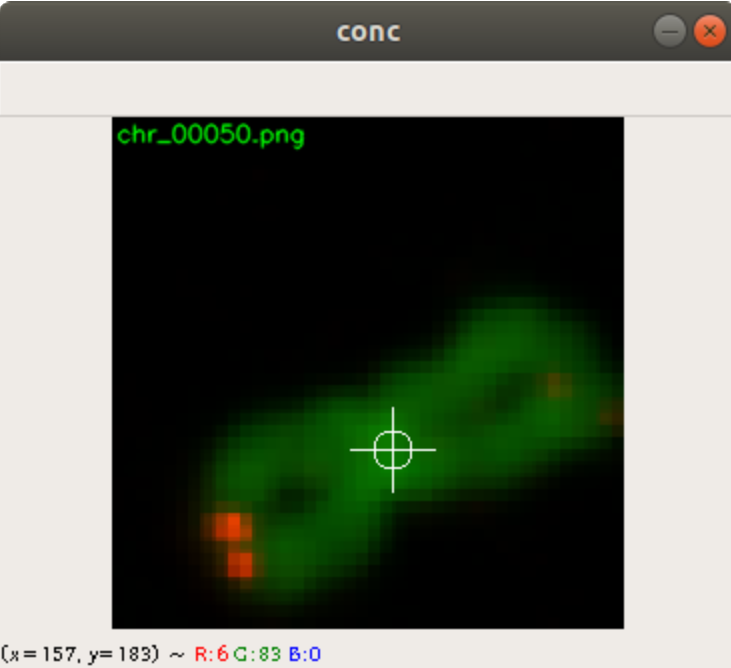
What is more, the background of the images from the database is extremely bright. This is not quite a real life background, which we find is much darker when you take pictures from inside a building. In Figure 2 you can see two different backgrounds which are surfaces from two different tables. The backgrounds do have relatively low brightness.
Cropping the images
The first task was to prepare the data for training the neural network. We decided to crop the images to the size of the fruits, so we receive some kind of standardization of the images. Below you find the code which crops the images to the size of the fruit. In this case we have the fruit images inside the addfolder. Inside the addfolder we first have two more directories, Testing and Training. Below these directories you find the directories for each fruit. We limit the number of fruits to six. The fruits we use are listed in dirlist, which are also the directory names.
The code is iterating through the Testing and Training directories and the fruit directories in dirlist and loads in every image with the opencv function imread. It converts the loaded image to a grayscale image and filters it with the opencv threshold function. After this we apply the findContours function which returns a list of contours of the image. The second largest contour (the largest contour has the size of the image itself) is taken and the width and height information of the contour is retrieved. The second largest contour is the fruit portion on the image. The application copies a square at the position of the second largest contour from the original image, resizes it to 100×100 pixels and saves it into a new directory destfolder.
srcfolder = '/home/inf/Bilder/Scale/orig/'
destfolder = '/home/inf/Bilder/Scale/cropped/'
addfolder = '/home/inf/Bilder/Scale/added/'
processedfolder = '/home/inf/Bilder/Scale/processed/'
dirtraintest = ['Testing', 'Training']
dirlist = ['Apfel','Gurke','Kartoffel','Orange','Tomate','Zwiebel']
count = 0
pattern = "*.jpg"
img_size = (100,100)
for traintest in dirtraintest:
for fruit in dirlist:
count = 0
for file in glob.glob(os.path.join(addfolder, traintest, fruit, pattern)):
im = cv2.imread(file)
imgray = cv2.cvtColor(im, cv2.COLOR_BGR2GRAY)
ret, thresh = cv2.threshold(imgray, 127, 200, 0)
contours, hierarchy = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
if len(contours) > 1:
cnt = sorted(contours, key=cv2.contourArea)
x, y, w, h = cv2.boundingRect(cnt[-2])
w = max((h, w))
h = w
crop_img = im[y:y+h, x:x+w]
im = cv2.resize(crop_img, img_size)
cv2.imwrite(os.path.join(destfolder, traintest, fruit, str("cropped_img_"+str(count)+".jpg")), im)
count += 1
Figure 1 shows how the application crops an image of an orange. On the left side, the orange fills out only part of the image. On the right side, the orange fills out the complete image.

Changing the backgrounds
Due to the extreme bright background of the images from the database we came to the decision to fill in new backgrounds on top of the bright ones. In Figure 2, you can see two different table surfaces, taken by the camera we used.

The code below shows how each image from the directory structure (which I explained above) is loaded into the variable pixels with the opencv imread function. Each pixel on each layer (RGB) of the image is checked, if a threshold of brightness has been reached. We assume that a pixel exceeding a certain brightness threshold is a background pixel (which is not always the case). The application then replaces the pixel with a pixel from a background image shown in Figure 2. It saves the new image to the directory processedfolder.
background = cv2.imread("background.jpg")
bg = np.zeros((img_size[0], img_size[1],3), np.uint8)
bgData = np.zeros((img_size[0], img_size[1],3), np.uint8)
bg = cv2.resize(background, img_size)
bgData = bg.copy()
threshold = (100, 100, 100)
for traintest in dirtraintest:
for fruit in dirlist:
count = 0
for name in glob.glob(os.path.join(destfolder, traintest, fruit, pattern)):
pixels = cv2.imread(os.path.join(destfolder, traintest, fruit, name))
pixelsData = pixels.copy()
for i in range(pixels.shape[0]): # for every pixel:
for j in range(pixels.shape[1]):
if pixelsData[i, j][0] >= threshold[0] and pixelsData[i, j][1] >= threshold[1] and pixelsData[i, j][2] >= threshold[2]:
pixelsData[i, j] = bgData[i, j]
cv2.imwrite(os.path.join(processedfolder, traintest, fruit, str("processed_img_"+str(count)+".jpg")), pixelsData)
count += 1
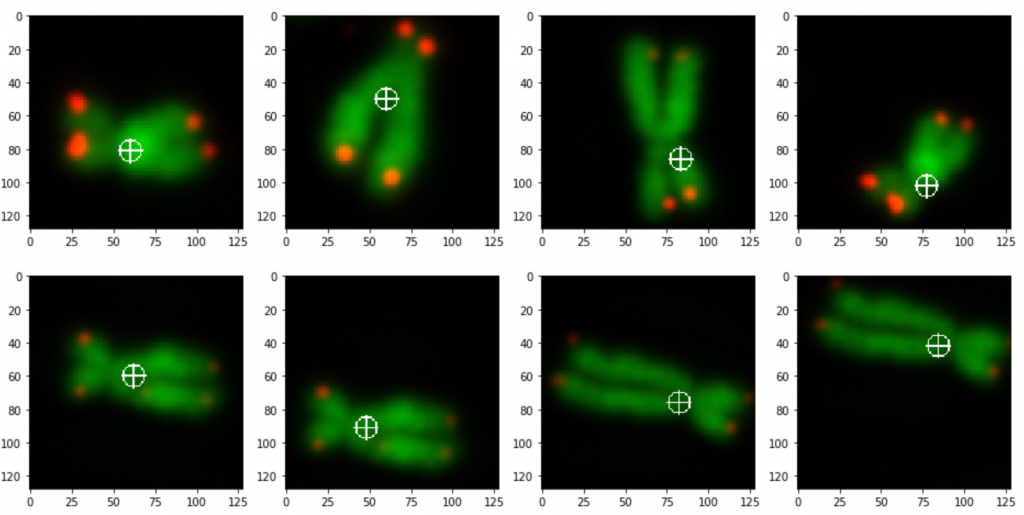
Figure 3 shows the output of two images from the code above. It shows the same orange with two different backgrounds.

Training the Model
Below the code of a neural network model. It consists of four convolutional layers. The number of filters is increased with each layer. After each convolutional layer there is a max pooling layer to reduce the image size for the input of the following layer. A flatten layer follows and is fed into a dense layer. Finally there is another dense layer with six neurons. This is the number of categories we have. Each layer uses the relu activation function. In the last layer however we use the softmax activation function. The reason for softmax, and not sigmoid, is, that we expect only one category from the six categories to be true for a given input image. This can be represented by the highest number calculated from the six output neurons. For optimization, we use stochastic gradient descent method.
model = Sequential() model.add(Conv2D(16, (3, 3), activation='relu', kernel_initializer='he_uniform', padding='same', input_shape=input_shape)) model.add(MaxPooling2D((2, 2))) model.add(Conv2D(32, (3, 3), activation='relu', kernel_initializer='he_uniform', padding='same')) model.add(MaxPooling2D((2, 2))) model.add(Dropout(0.1)) model.add(Conv2D(64, (3, 3), activation='relu', kernel_initializer='he_uniform', padding='same')) model.add(MaxPooling2D((2, 2))) model.add(Dropout(0.1)) model.add(Conv2D(128, (3, 3), activation='relu', kernel_initializer='he_uniform', padding='same')) model.add(MaxPooling2D((2, 2))) model.add(Dropout(0.1)) model.add(Flatten()) model.add(Dense(256, activation='relu', kernel_initializer='he_uniform')) model.add(Dropout(0.1)) model.add(Dense(6, activation='softmax')) opt = SGD(lr=0.001, momentum=0.9) model.compile(optimizer=opt, loss='categorical_crossentropy', metrics=['accuracy'])
We load in all training and validation images from the directory train_path and valid_path with the Keras ImageDataGenerator. By doing this the ImageDataGenerator rescales the images and augment the images by shifting and flipping. The training and validation images from the directories train_path and valid_path are moved into the lists train_it and valid_it. The method flow_from_directory makes this task easy since it considers the directory structure below the directories train_path and valid_path, as well. In our case, we have the directories Apfel, Gurke, Kartoffel, Orange, Tomate, Zwiebel below of train_path and valid_path. In each of these directories you find the corresponding images (such all apple images in directory Apfel, all cucumber images in directory Gurke etc.).
train_datagen = ImageDataGenerator(rescale=1.0/255.0,width_shift_range=0.1, height_shift_range=0.1, horizontal_flip=True) test_datagen = ImageDataGenerator(rescale=1.0/255.0) train_it = train_datagen.flow_from_directory(train_path,class_mode='categorical', batch_size=64, target_size=image_size) valid_it = test_datagen.flow_from_directory(valid_path,class_mode='categorical', batch_size=64, target_size=image_size)
The training is started with the Keras fit_generator command. It uses the lists train_it and valid_it as inputs. We defined a callback function to produce checkpoints from the neural network weights, each time the training shows some improvement concerning validation loss.
callbacks = [
EarlyStopping(patience=10, verbose=1),
ReduceLROnPlateau(factor=0.1, patience=3, min_lr=0.00001, verbose=1),
ModelCheckpoint('modelmulticat.h5', verbose=1, save_best_only=True, save_weights_only=True)
]
history = model.fit_generator(train_it, steps_per_epoch=len(train_it),validation_data=valid_it, validation_steps=len(valid_it), epochs=10, callbacks=callbacks, verbose=1)
_, acc = model.evaluate_generator(valid_it, steps=len(valid_it), verbose=0)
print('> %.3f' % (acc * 100.0))
model_json = model.to_json()
with open("modelmulticat.json", "w") as json_file:
json_file.write(model_json)
Finally the structure of the trained model is saved to a json file.
The training time with this model is about three minutes on a NVIDIA graphics card. We use about 6000 images for training and 2000 images for validation, altogether. The validation accuracy was 96% which was above the accuracy, which shows a little underfitting.
Testing the Model
We tested the model with the code below. First, we loaded the image in the variable img with the opencv function imread read. Right after this, we have to take care of the image layers. The way opencv handles the image layers is different from the way Keras with its predict method does. They have the Red and the Blue layers switched. For this reason, we have to apply the cvtColor method, which switches the Red and Blue layers. The image is then normalized by dividing its pixels values with 255. Finally the prediction method is used to predict the image. Figure 4 shows an example of an image for input, which is printed out by the matplotlib function imshow. The method predict returns a probability vector predictions. The index with the highest value of the vector corresponds to the category. The category can be retrieved from the class_indices list.
img = cv2.imread(os.path.join(valid_path,"Apfel/cropped_img_592.jpg"),1) img = cv2.cvtColor(img, cv2.COLOR_BGR2RGB) imshow(img) img = np.array(img, dtype=np.float32) img *= 1.0/255.0 predictions = model.predict([[img]]) print(predictions) result = np.where(predictions[0] == np.amax(predictions[0])) assert len(result)==1 print(list(valid_it.class_indices)[result[0][0]])
We tested a few times with different image and saw that the prediction delivered pretty good results.

The Raspberry Pi application
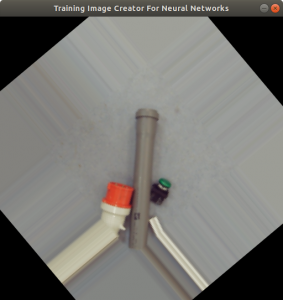
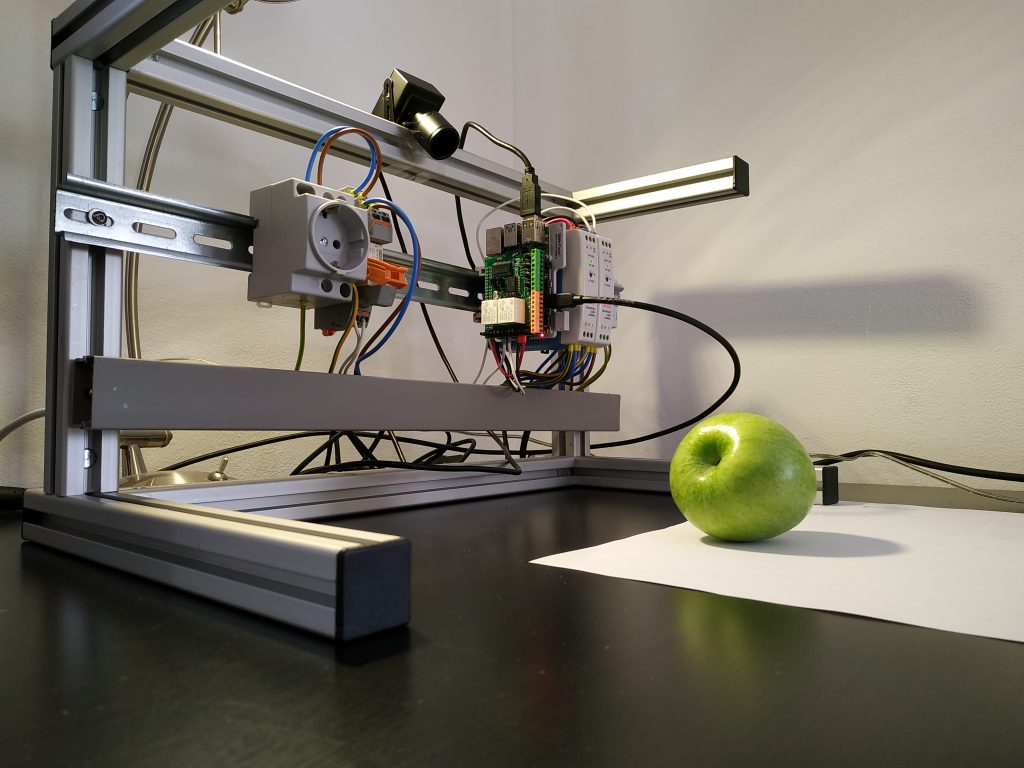
The setup of the experiment is shown in Figure 5. The raspberry pi 4, power supply and a socket are mounted on a top-hat rail. On the raspberry pi you see a piface shield attached. The shield had to be mechanically prepared to fit on a raspberry pi 4. The shield provides buttons in case it is needed. Additionally we have a relay and a power socket. The relay can be triggered by the piface, so the relay applies 230V to the socket. On top of the construction you find an usb camera.
We defined a function getCrop, see code below, which crops the image to the size of the portion of the fruit. This procedure was already explained above. Here we introduced the variable threshset, where the user can modify the threshold value of the opencv threshold method using keys. This is explained later.
threshset = 100
def getCrop(im):
global threshset
imgray = cv2.cvtColor(im, cv2.COLOR_BGR2GRAY)
ret, thresh = cv2.threshold(imgray, threshset, 255, 0)
contours, hierarchy = cv2.findContours(thresh, cv2.RETR_TREE, cv2.CHAIN_APPROX_SIMPLE)
if len(contours) >= 1:
cnts = sorted(contours, key=cv2.contourArea, reverse=True)
for cnt in cnts:
x, y, w, h = cv2.boundingRect(cnt)
if w > im.shape[0]*20//100 and w < im.shape[0]*95//100:
if h > im.shape[1]*20//100 and h < im.shape[1]*95//100:
w = max((h, w))
h = w
return x,y,w
return 0,0,0
In the beginning we faced the problem that the neural network did not predict very well due to too few training images. Therefore we introduced a function to save easily badly predicted images. The name of the function is saveimg. It simply saves an image img to a directory with a name containing the parameters dircat and fruit. The image name also contains the date and the time.
def saveimg(img, dircat, fruit):
global croppedfolder
now = datetime.now()
dt_string = now.strftime("%d_%m_%Y_%H_%M_%S")
resized = np.zeros((image_size[0], image_size[1],3), np.uint8)
resized = cv2.resize(img, image_size, interpolation = cv2.INTER_AREA)
cv2.imwrite(os.path.join(croppedfolder, dircat, fruit, str("img_"+dt_string+".jpg")), resized)
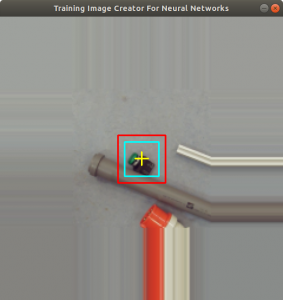
Below you find the raspberry pi application code. In the beginning it sets up the opencv video feature. Inside the while loop, an image frame from the usb camera is taken, which is then copied into the image objectfr. The function getCrop is used to get the fruit portion of the image and a rectangle is drawn around the fruit portion. The function putText writes the current value of threshset into the image objectfr as well. The application then shows the modified image on a display, see Figure 6. The opencv method waitkey checks for a pressed key. In case a key was pressed, code depending on the key will be executed.
cam = cv2.VideoCapture(0)
cv2.namedWindow("object")
while True:
ret, frame = cam.read()
if not ret:
print("cam.read something wrong")
break
objectfr = frame.copy()
x,y,w = getCrop(objectfr)
cv2.rectangle(objectfr, (x,y), (x+w,y+w), (0,255,0), 1)
cv2.putText(objectfr, "thresh: {}".format(threshset), (10,30), cv2.FONT_HERSHEY_SIMPLEX, 1, (0, 255, 0), 1, cv2.LINE_AA)
cv2.imshow("object", objectfr)
if not ret:
break
k = cv2.waitKey(1)
if k & 0xFF == ord('q') :
break
elif k & 0xFF == ord('n') :
resized = np.zeros((image_size[0], image_size[1],3), np.uint8)
resized = cv2.resize(frame[y:y+w,x:x+w,:], image_size, interpolation = cv2.INTER_AREA)
cv2.imwrite("checkpic.jpg",resized)
resized = cv2.cvtColor(resized, cv2.COLOR_BGR2RGB)
resized = np.array(resized, dtype=np.float32)
resized *= 1.0/255.0
predictions = model.predict([[resized]])
print(predictions)
result = np.where(predictions[0] == np.amax(predictions[0]))
assert len(result)==1
print(result[0][0])
print(list(valid_it.class_indices)[result[0][0]])
os.system("espeak -vde {}".format(list(valid_it.class_indices)[result[0][0]]))
elif k & 0xFF == ord('a'):
saveimg(frame[y:y+w,x:x+w,:], "Training", "Apfel")
img_counter += 1
elif k & 0xFF == ord('z'):
saveimg(frame[y:y+w,x:x+w,:], "Training", "Zwiebel")
img_counter += 1
elif k & 0xFF == ord('o'):
saveimg(frame[y:y+w,x:x+w,:], "Training", "Orange")
img_counter += 1
elif k & 0xFF == ord('k'):
saveimg(frame[y:y+w,x:x+w,:], "Training", "Kartoffel")
img_counter += 1
elif k & 0xFF == ord('+'):
threshset += 5
if threshset > 255:
threshset = 255
elif k & 0xFF == ord('-'):
threshset -= 5
if threshset < 0:
threshset = 0
cam.release()
cv2.destroyAllWindows()
If the key ‘q’ is pressed, than the application stops. If the key ‘n’ is pressed, the image inside the rectangle is taken and the category is predicted with the Keras predict method. The string is handed over to the espeak application which speaks out the category on the speaker attached on the raspberry pi. The keys ‘a’, ‘z’, ‘o’, ‘k’ execute the saveimg function with different parameters. The purpose of these keys is, that the user can save an image, in case there is a bad prediction. Next time, the model is trained, the saved image will be included in the training data. At last we have the ‘+’ and ‘-‘ keys, which modify the threshset value. The effect will be, that the rectangle (Figure 6, green rectangle) is enlarged or downsized due to the shadow on the background.

Conclusion
The application works amazingly well with few fruits to predict considering the relative low number of training data. In the beginning we had to retrain the model a couple of times with newly generated images using the application keys described above.
As soon as we take e.g. an apple with different colors, there is a high chance that the prediction fails. In such cases we have take more images and retrain again.
Acknowledgement
Thanks to Carmen Furch and Armin Weisser providing the data preparation code and the raspberry pi application.
Also special thanks to the University of Applied Science Albstadt-Sigmaringen offering a classroom and appliances to enable this research.